Binding Sites Prediction of TetR Family Repressors
This server can be used to predict DNA binding sites for transcription factors of TetR Family Repressors (TFRs). Two methods as described in the reference at the end of this page are used: a genome sequence-based method which uses ideas of phylogenetic footprinting, and a statistical energy-based method which calculates sequence energies of DNA octamers given the amino acid sequences of TFRs. Without user-provided DNA sequences, TFBSs are predicted as genome sequence fragments near the ORF of a query TFR, and as octamer DNA sequences and sequence logos representing predicted half binding sites of a query TFR. If a DNA sequence is provided by user, the given DNA sequence will be scanned for most possible TFBSs.
For detailed descripstion of the method, please read the references.
The source code and demos can be downloaded here.
After submission, the calculation will be carried out on our servers. You will be navigated to a page waiting for it to finish. That page will be automatically updated with downloadable results after the calculation is finished.
References:
- Long, Pengpeng; Zhang, Lu; Huang, Bin; Chen, Quan*; Haiyan, Liu*; Integrating genome sequence and structural data for statistical learning to predict transcription factor binding sites, Nucleic Acids Research, 2020, 48(22): 12604-12617.
ABACUS2: Protein Sequence Design
To design amino acid sequences that are intended to fold into a user-provided backbone structure. This is achieved by optimizing the ABACUS (A Backbone-based Amino aCid Usage Survey) statistical energy function with respect to the sequence.
For detailed descriptions of the method, please read the references.
If you only need to score the compatibility between an existing sequence and a backbone structure using the ABACUS model, you may use the ABACUS Score service.
After submission, the calculation will be carried out on our servers. You will be navigated to a page waiting for it to finish. That page will be automatically updated with downloadable results after the calculation is finished.
If you have any problem or suggestion, please contact <yuhh@mail.ustc.edu.cn>.
References:
- Xiong P, Wang M, et al. Protein design with a comprehensive statistical energy function and boosted by experimental selection for foldability[J]. Nature communications, 2014, 5: 5330.
- Xiong P, et al. Increasing the Efficiency and Accuracy of the ABACUS Protein Sequence Design Method[J]. Bioinformatics, 2020, 36(1): 136-144.
ABACUS2: Score Protein Sequence for Given Structure
To score the compatibility between an amino acid sequence and a backbone structure contained in the user-provided input PDB file. The ABACUS (A Backbone-based Amino aCid Usage Survey) sequence energy will be evaluated. The returned results will contain energy contributions of different energy components at different amino acid positions.
If you have any problem or suggestion, please contact us
References:
- Xiong P, Wang M, et al. Protein design with a comprehensive statistical energy function and boosted by experimental selection for foldability[J]. Nature communications, 2014, 5: 5330.
- Xiong P, et al. Increasing the Efficiency and Accuracy of the ABACUS Protein Sequence Design Method[J]. Bioinformatics, 2020, 36(1): 136-144.
SCUBA Download Page
What is SCUBA?
SCUBA (SideChain Unspecialized Backbone Arrangement) is a statistical energy function of protein conformation. It consists of energy terms derived from known protein structures using a novel adaptive-kernel neighbor counting-neural network(NC-NN) approach. It is continuous with analytical gradients, allowing protein structures to be sampled and/or optimized with complete flexibility by stochastics dynamics (SD) simulations.
What can SCUBA be used for?
By design, SCUBA energy contains both local and through-space packing interactions of mainchain atoms, while sidechains in the model mainly serve as steric placeholders. In SCUBA-driven protein backbone design, SD simulated annealing can be applied to generate optimized backbone structures at high resolution from an initial backbone which can be partially or entirely artificially constructed. During the optimization, generic instead of specific sidechain types can be employed, solving the problem of designing backbones without knowing the amino acid sequence in advance.
What are included in the downloaded package?
- Statically linked binaries to run SCUBA-SD on x86_64 linux machines.
- Documented scripts for the user to setup and run the SCUBA-SD of their own protein systems.
- Demos illustrating how SCUBA-SD is used. The first is to optimize an entirely artificially constructed backbone. The second is to optimize a backbone with artificially constructed parts. The last is to sample a native protein structure around its native conformation.
Examples of SCUBA-SD Optimized Backbones
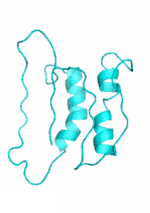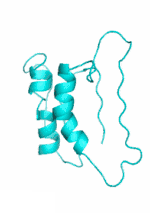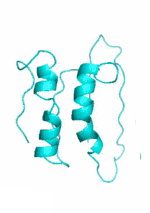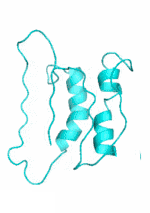
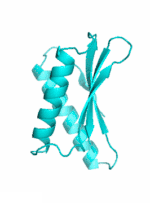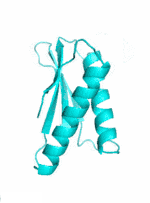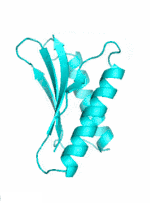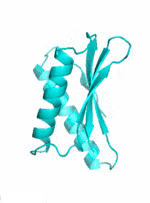
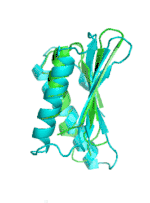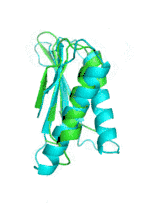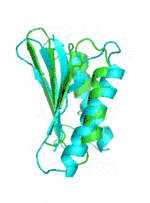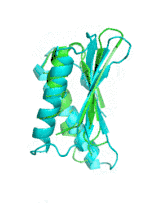
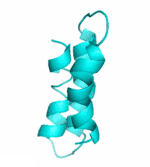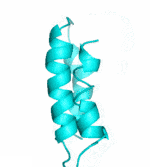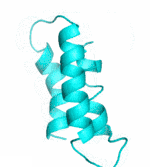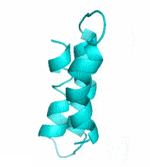
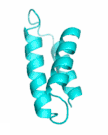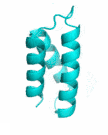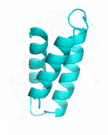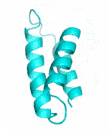

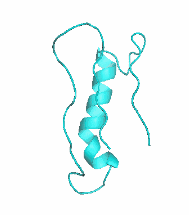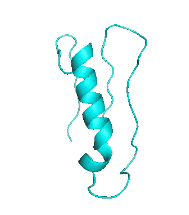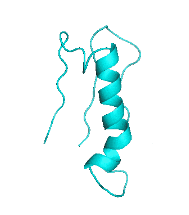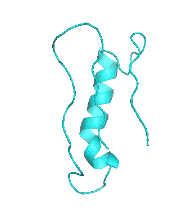
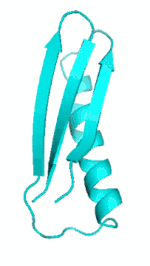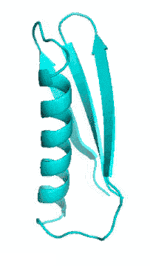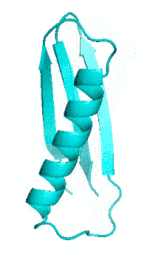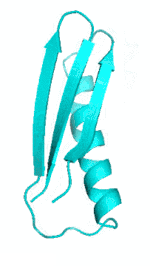
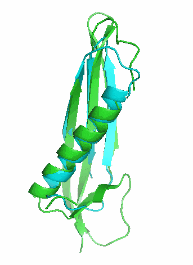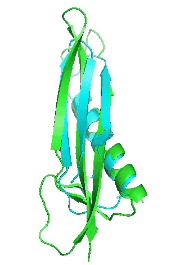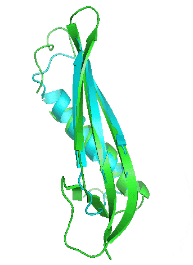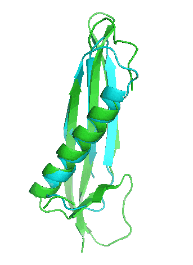
References:
- Bin Huang#, Yang Xu#, Xiuhong Hu#, Yongrui Liu, Shanhui Liao, Jiahai Zhang, Chengdong Huang, Jingjun Hong, Quan Chen*, Haiyan Liu*, A backbone-centred energy function of neural networks for protein design, Nature, 602: 523–528 (2022)
Download Other
structure_5816.tar.gz contain 5816 designed prtotein structures contain cavity.
References:
- De novo Design of Cavity-containing Proteins with a Backbone-centred Network Energy Function.