| Servers |
| Downloads |
SCUBA Download Page
What is SCUBA? reference
SCUBA (SideChain Unknown Backbone Arrangement) is a statistical energy function of protein conformation. It consists of energy terms derived from known protein structures using a novel adaptive-kernel neighbor counting-neural network(NC-NN) approach. It is continuous with analytical gradients, allowing protein structures to be sampled and/or optimized with complete flexibility by stochastics dynamics (SD) simulations.
What can SCUBA be used for?
By design, SCUBA energy contains both local and through-space packing interactions of mainchain atoms, while sidechains in the model mainly serve as steric placeholders. In SCUBA-driven protein backbone design, SD simulated annealing can be applied to generate optimized backbone structures at high resolution from an initial backbone which can be partially or entirely artificially constructed. During the optimization, generic instead of specific sidechain types can be employed, solving the problem of designing backbones without knowing the amino acid sequence in advance.
What are included in the downloaded package?
1. Statically linked binaries to run SCUBA-SD on x86_64 linux machines.
2. Documented scripts for the user to setup and run the SCUBA-SD of their own protein systems.
3. Demos illustrating how SCUBA-SD is used. The first is to optimize an entirely artificially constructed backbone. The second is to optimize a backbone with artificially constructed parts. The last is to sample a native protein structure around its native conformation.
| Examples | Arbitrarily Constructed Initial Backbone | SCUBA-SD Optimized Backbone | SCUBA-SD Optimized Backbone Matching with Native Backbone |
| I | 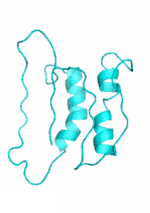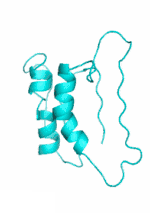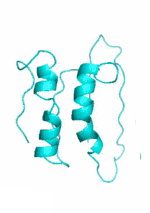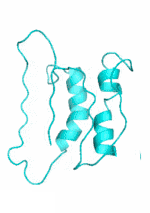 |
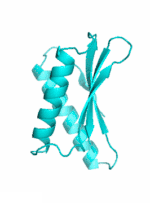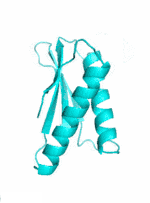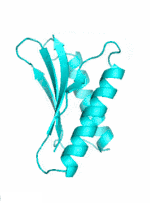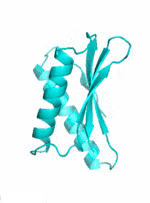 |
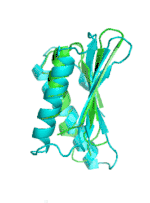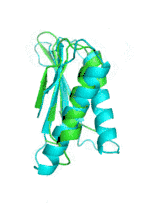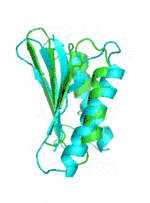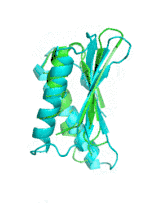 |
| II | 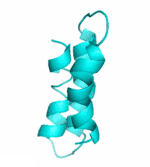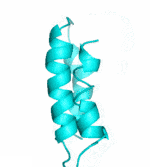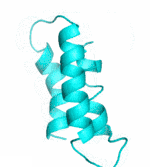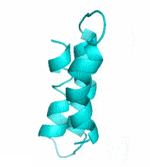 |
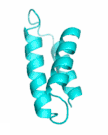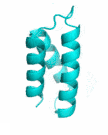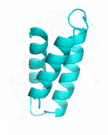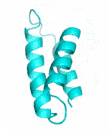 |
 |
| III | 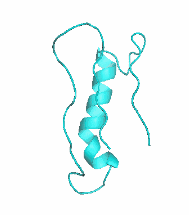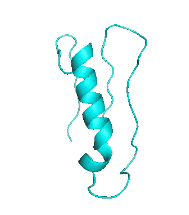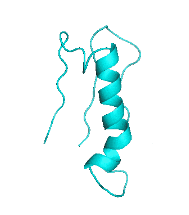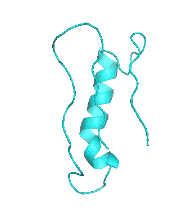 |
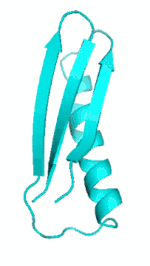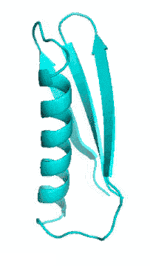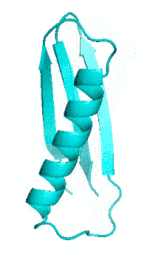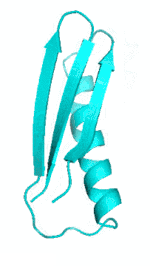 |
 |
If you have any problem or suggestion, please contact <hb080@mail.ustc.edu.cn>.
Reference:
- Bin Huang, Yang Xu, Xiuhong Hu, Yongrui Liu, Shanhui Liao, Jiahai Zhang, Chengdong Huang, Jingjun Hong, Quan Chen*, Haiyan Liu*, A backbone-centred energy function of neural networks for protein design. Nature, 2022, 602(7987):523-528. doi:10.1038/s41586-021-04383-5.